Next to be submitted at Oxford Bioinformatics 2023
BioQAGNN
Reasoning with Language Models and Knowledge Graphs for Biomedical Open-Domain Question Answering
Giacomo Frisoni, Enrico Gnagnarella, Luca Ragazzi, Gianluca Moro, Antonella Carbonaro
Description
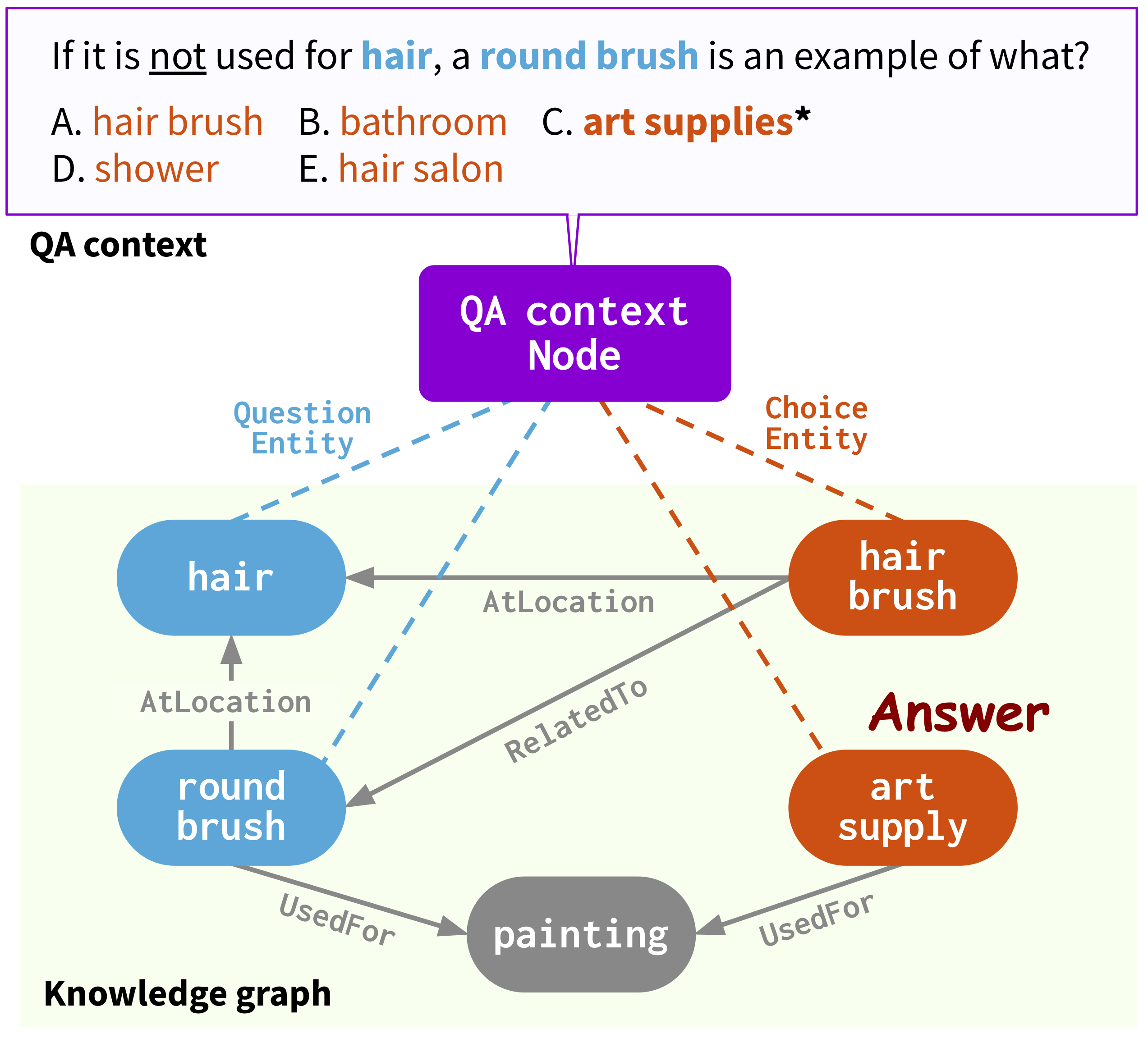
Injecting world or domain-specific structured knowledge into pre-trained language models (PLMs) is becoming an increasingly popular approach for solving problems such as biases, hallucinations, huge architectural sizes, and explainability lack—critical for real-world natural language processing applications in sensitive fields like bioinformatics. One recent work that has garnered much attention in Neuro-symbolic AI is QA-GNN by Yasunaga et al. (2021), an end-to-end model for multiple-choice open-domain question answering (MCOQA) tasks via interpretable text-graph reasoning. Unlike previous publications, QA-GNN mutually informs PLMs and graph neural networks (GNNs) on top of relevant facts retrieved from knowledge graphs (KGs). However, taking a more holistic view, existing PLM+KG contributions mainly consider commonsense benchmarks and ignore or shallowly analyze performances on biomedical datasets. This paper proposes a deep investigation of QA-GNN for biomedicine, comparing existing or brand-new PLMs, KGs, edge-aware GNNs, preprocessing techniques, and initialization strategies. By combining the insights emerged in our study, we introduce Bio-QA-GNN, a new state-of-the-art MCOQA model on biomedical/clinical text, largely outperforming the original one. Our findings also contribute to a better understanding of the explanation degree allowed by joint text-graph reasoning architectures and their effectiveness on different medical subjects and reasoning types.
Keywords: open-domain question answering, biomedical natural language processing, neuro-symbolic, subgraph retrieval, graph neural network.